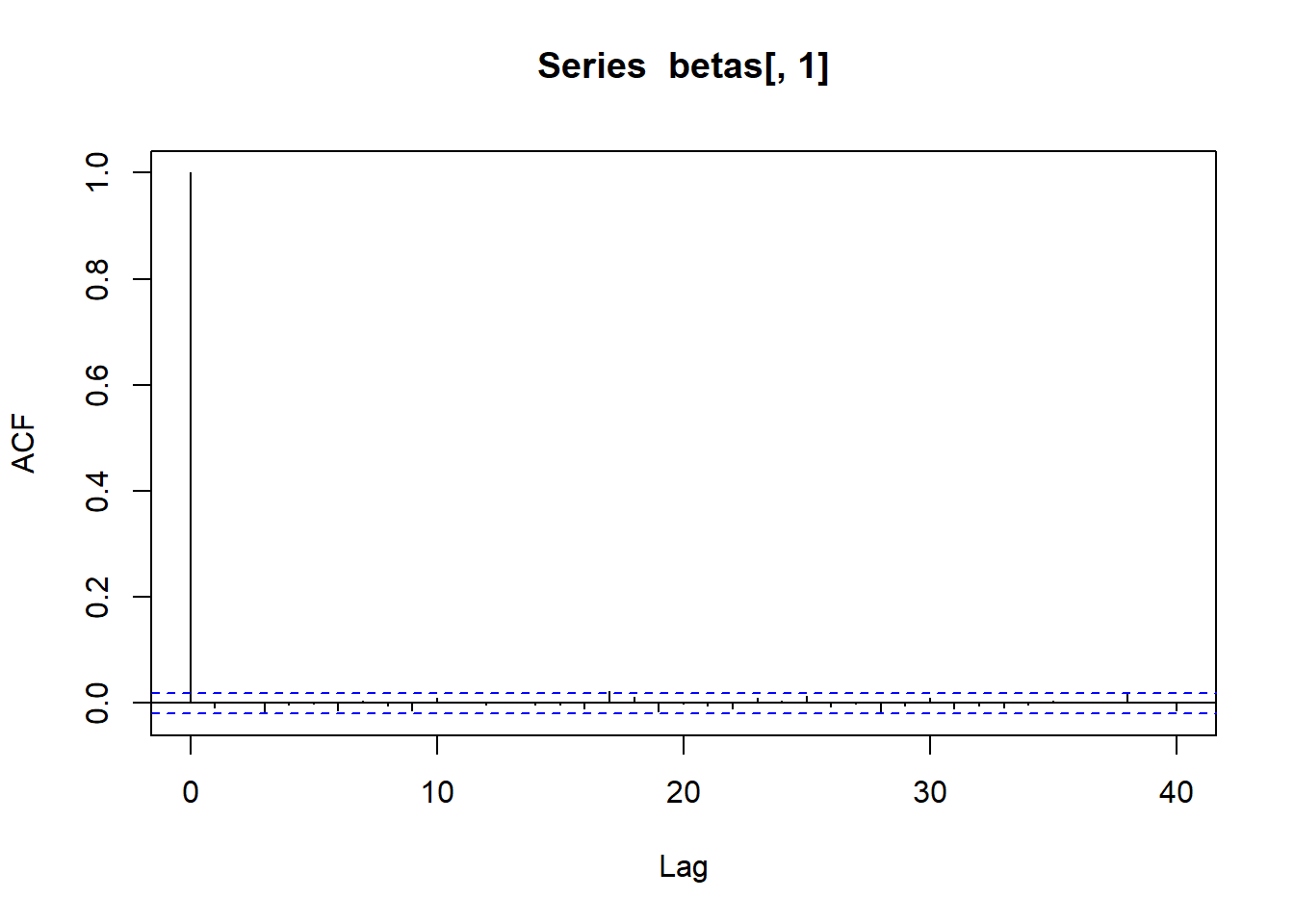You can download the R script here .
Some useful links for working with Stan:
Linear Regression
In this part, we are going to fit a simple linear regression model through Stan. Here you can download the .stan model.
First of all, let’s compile the model and save it into an R object.
<- rstan:: stan_model (file= "data/LM_Model.stan" )
S4 class stanmodel 'anon_model' coded as follows:
data{
int<lower = 0> n; // number of obs
int<lower = 0> K; // number of covariates (including the intercept)
vector[n] y; // response
matrix[n, K] X; // covariates
real a0;
real b0;
vector[K] beta0;
vector[K] s2_0;
}
parameters{
real<lower = 0> sigma2;
vector[K] beta;
}
transformed parameters {
vector[n] mu;
mu = X * beta;
}
model{
// Prior:
sigma2 ~ inv_gamma(a0, b0);
for(k in 1:K){
beta[k] ~ normal(beta0[k], sqrt(s2_0[k]));
}
// Likelihood:
y ~ normal(mu, sqrt(sigma2));
}
generated quantities{
vector[n] log_lik;
for (j in 1:n){
log_lik[j] = normal_lpdf(y[j] | mu[j], sqrt(sigma2));
}
}
We consider the Prestige data from the car package. Statistical units are occupations, for which we have:
education: average education of occupational incumbents, years, in 1971;
income: average income of incumbents, dollars, in 1971;
women: percentage of incumbents who are women;
prestige: Pineo-Porter prestige score for occupation, from a social survey conducted in the mid-1960s;
census: canadian Census occupational code;
type: type of occupation (bc, Blue Collar; prof, Professional, Managerial, and Technical; wc, White Collar).
Let’s transform the income variable so to reduce the asimmetry.
data ("Prestige" )<- complete.cases (Prestige),- 5 ]$ income <- log (Prestige2$ income)
In order to fit the stan model, we need to pass some data: - y: the response vector;
X: the design matrix;
n: the sample size;
K: the number of covariates (including the intercept term);
a0, b0, beta0, s2_0: hyperparameters.
<- Prestige2$ prestige<- lm (prestige ~ ., data= Prestige2)<- model.matrix (mod)<- nrow (X)<- ncol (X)
We further create a list containing the objects to be passed to the stan model.
<- list (y = y,X = X,n = n,K = K,a0 = 10 ,b0 = 10 ,beta0 = rep (0 , K),s2_0 = rep (10 , K)
We can now use the rstan::sampling function to fit the model. We what two chains, each of length 10’000, and we want to discard the first 50% as warm-up.
<- 10000 <- 2 <- rstan:: sampling (object = LM_Model,data = data.stan,warmup = 0.5 * n.iter, iter = n.iter,thin= 1 , chains = nchain,refresh = n.iter/ 2 #, pars=c("beta", "sigma2")
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 7.1e-05 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.71 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 10000 [ 0%] (Warmup)
Chain 1: Iteration: 5000 / 10000 [ 50%] (Warmup)
Chain 1: Iteration: 5001 / 10000 [ 50%] (Sampling)
Chain 1: Iteration: 10000 / 10000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 1.551 seconds (Warm-up)
Chain 1: 1.887 seconds (Sampling)
Chain 1: 3.438 seconds (Total)
Chain 1:
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 2).
Chain 2:
Chain 2: Gradient evaluation took 1.3e-05 seconds
Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.13 seconds.
Chain 2: Adjust your expectations accordingly!
Chain 2:
Chain 2:
Chain 2: Iteration: 1 / 10000 [ 0%] (Warmup)
Chain 2: Iteration: 5000 / 10000 [ 50%] (Warmup)
Chain 2: Iteration: 5001 / 10000 [ 50%] (Sampling)
Chain 2: Iteration: 10000 / 10000 [100%] (Sampling)
Chain 2:
Chain 2: Elapsed Time: 1.666 seconds (Warm-up)
Chain 2: 1.78 seconds (Sampling)
Chain 2: 3.446 seconds (Total)
Chain 2:
<- summary (LM_stan, pars = c ("beta" , "sigma2" ))$ summaryrownames (summ)[1 : K] <- colnames (X)round (summ, 3 )
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff
(Intercept) -2.968 0.041 3.166 -9.251 -5.121 -2.989 -0.828 3.335 6073.251
education 4.306 0.007 0.452 3.431 3.997 4.309 4.608 5.183 4014.248
income 0.507 0.010 0.618 -0.689 0.081 0.505 0.925 1.712 4123.754
women -0.060 0.000 0.025 -0.108 -0.076 -0.060 -0.044 -0.012 6170.313
typeprof 6.119 0.034 2.255 1.602 4.600 6.131 7.615 10.485 4498.555
typewc -2.776 0.024 1.791 -6.340 -3.971 -2.789 -1.575 0.760 5570.694
sigma2 48.765 0.079 6.868 37.081 43.948 48.202 52.919 63.777 7574.694
Rhat
(Intercept) 1
education 1
income 1
women 1
typeprof 1
typewc 1
sigma2 1
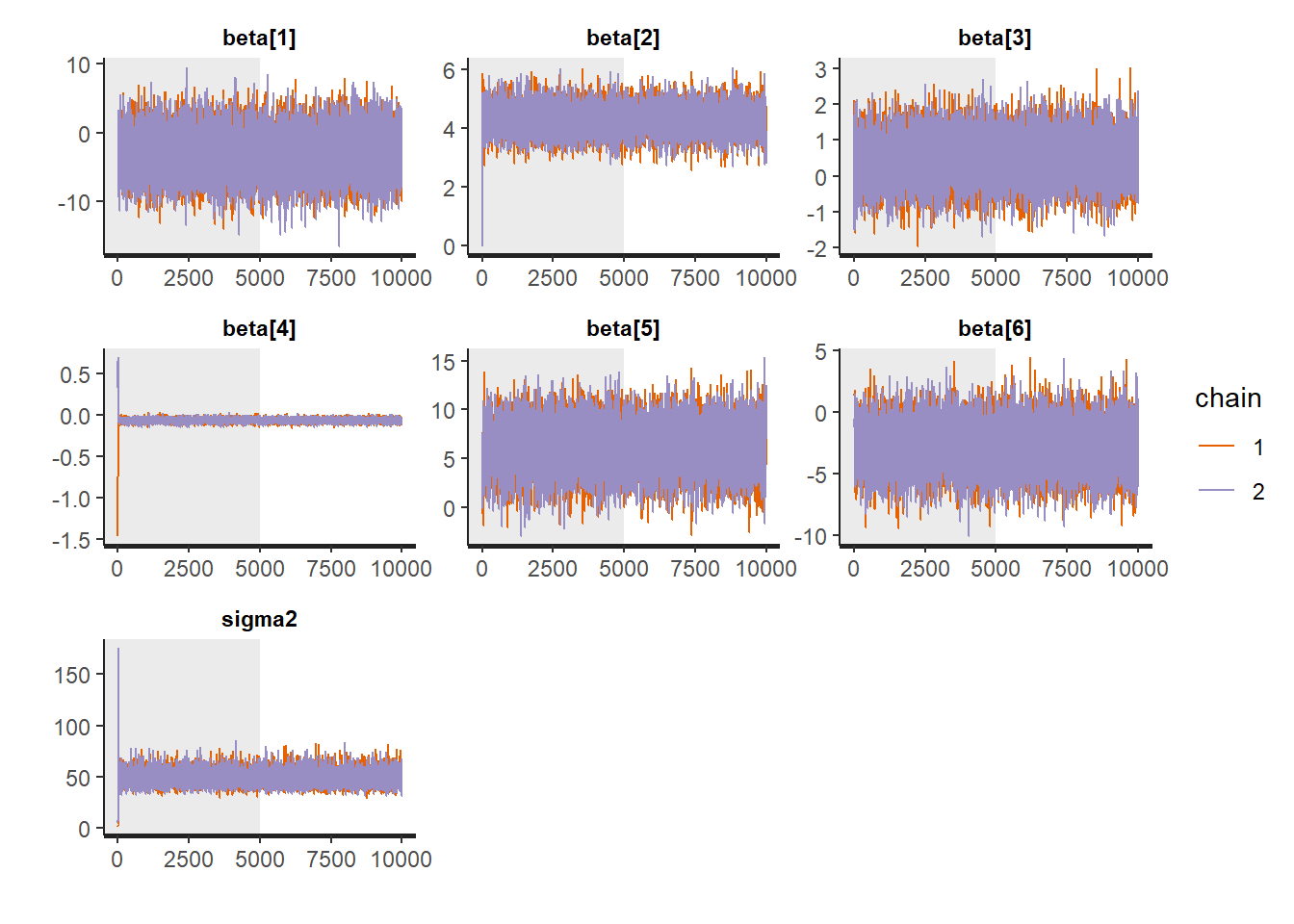
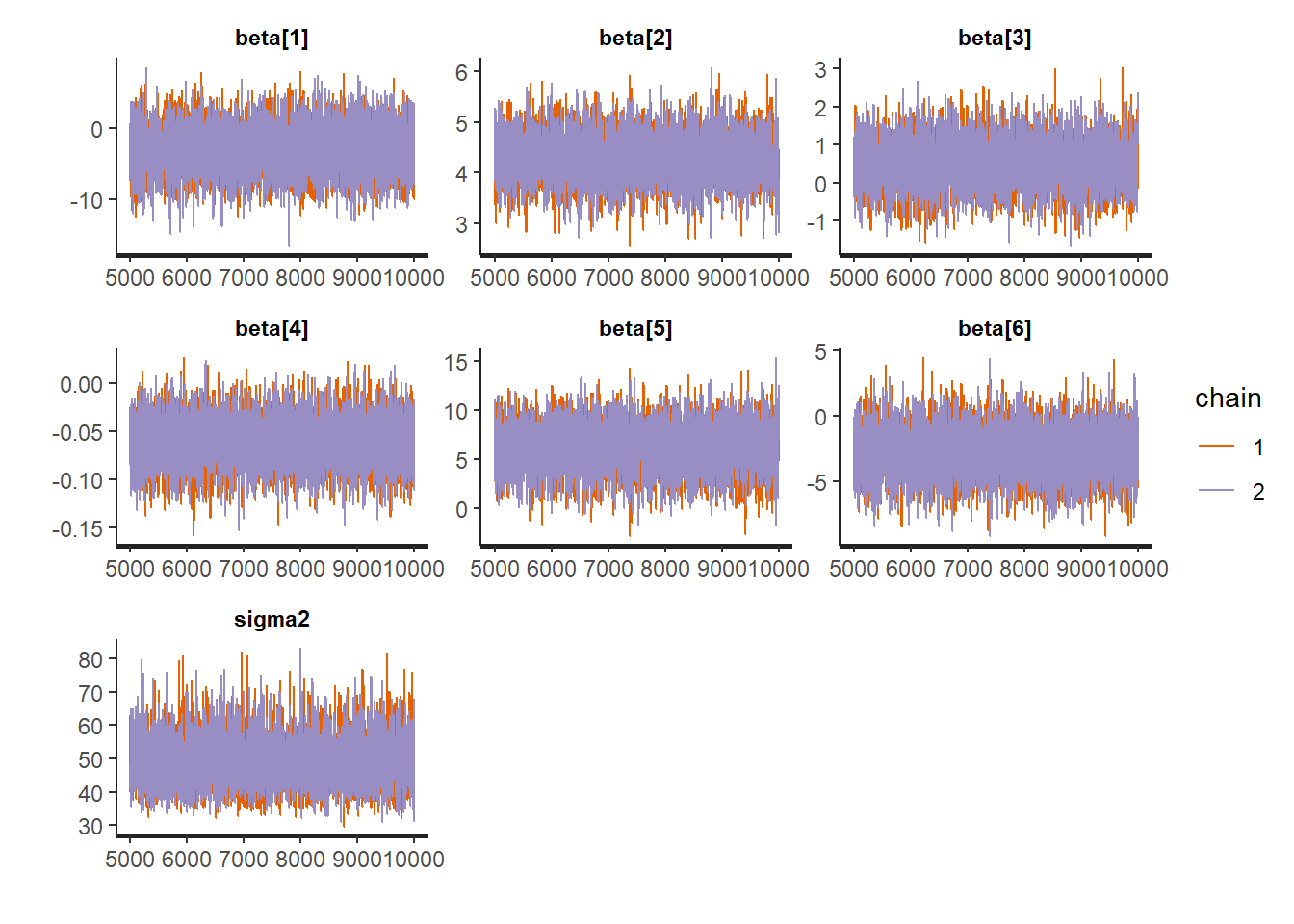
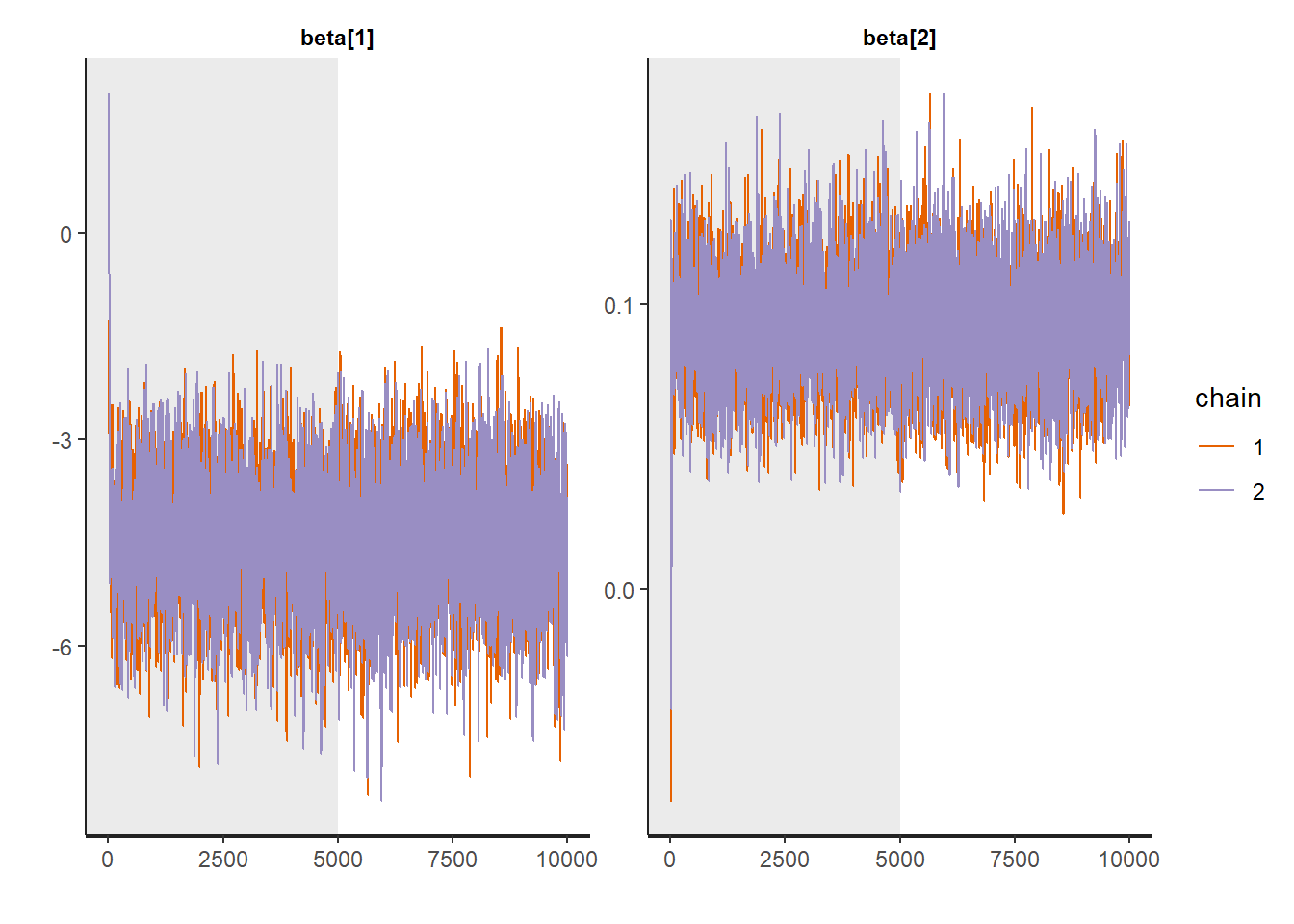
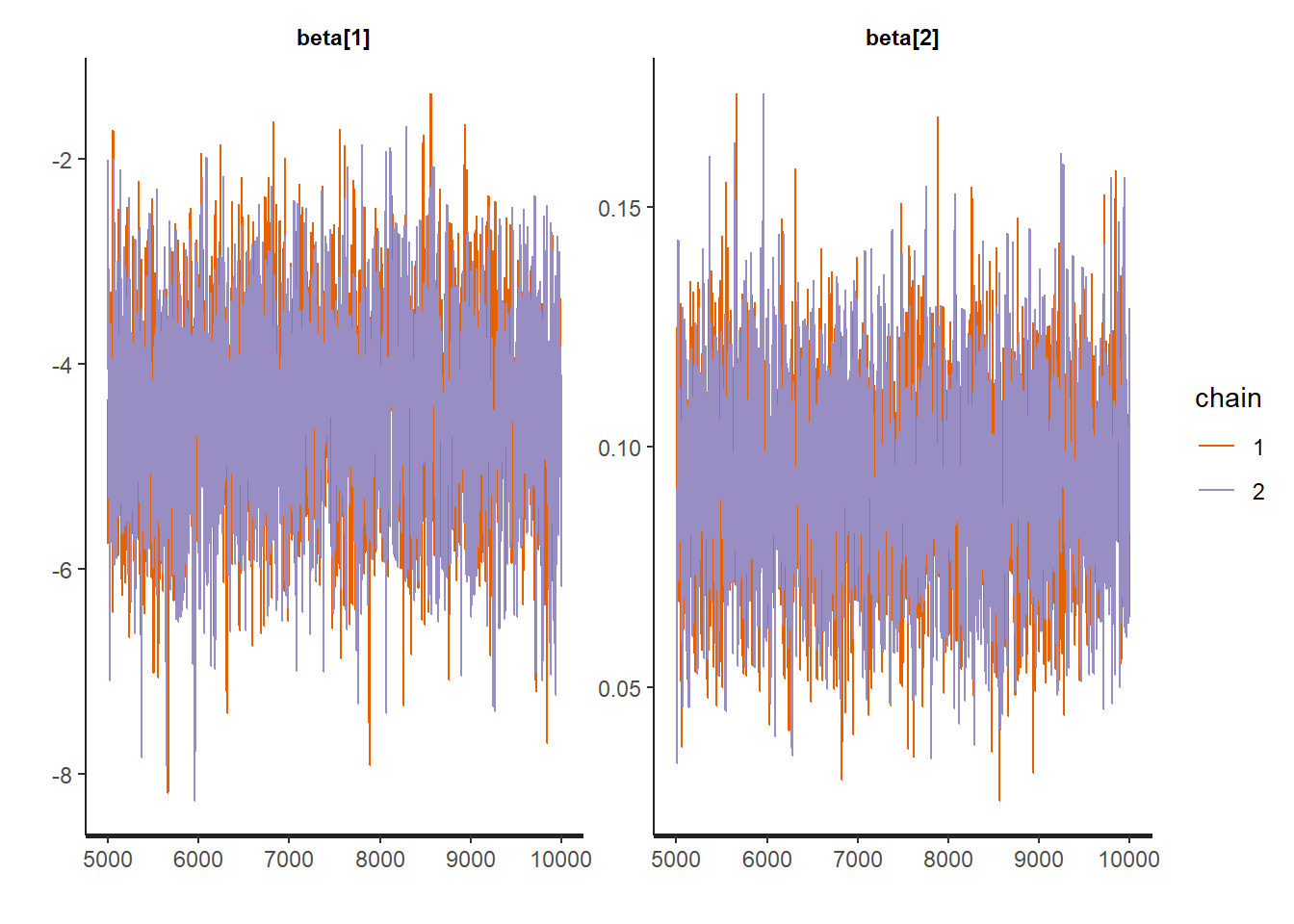
Traceplots:
:: traceplot (LM_stan, pars = c ('beta' , 'sigma2' ), inc_warmup = TRUE ):: traceplot (LM_stan, pars = c ('beta' , 'sigma2' ), inc_warmup = FALSE )
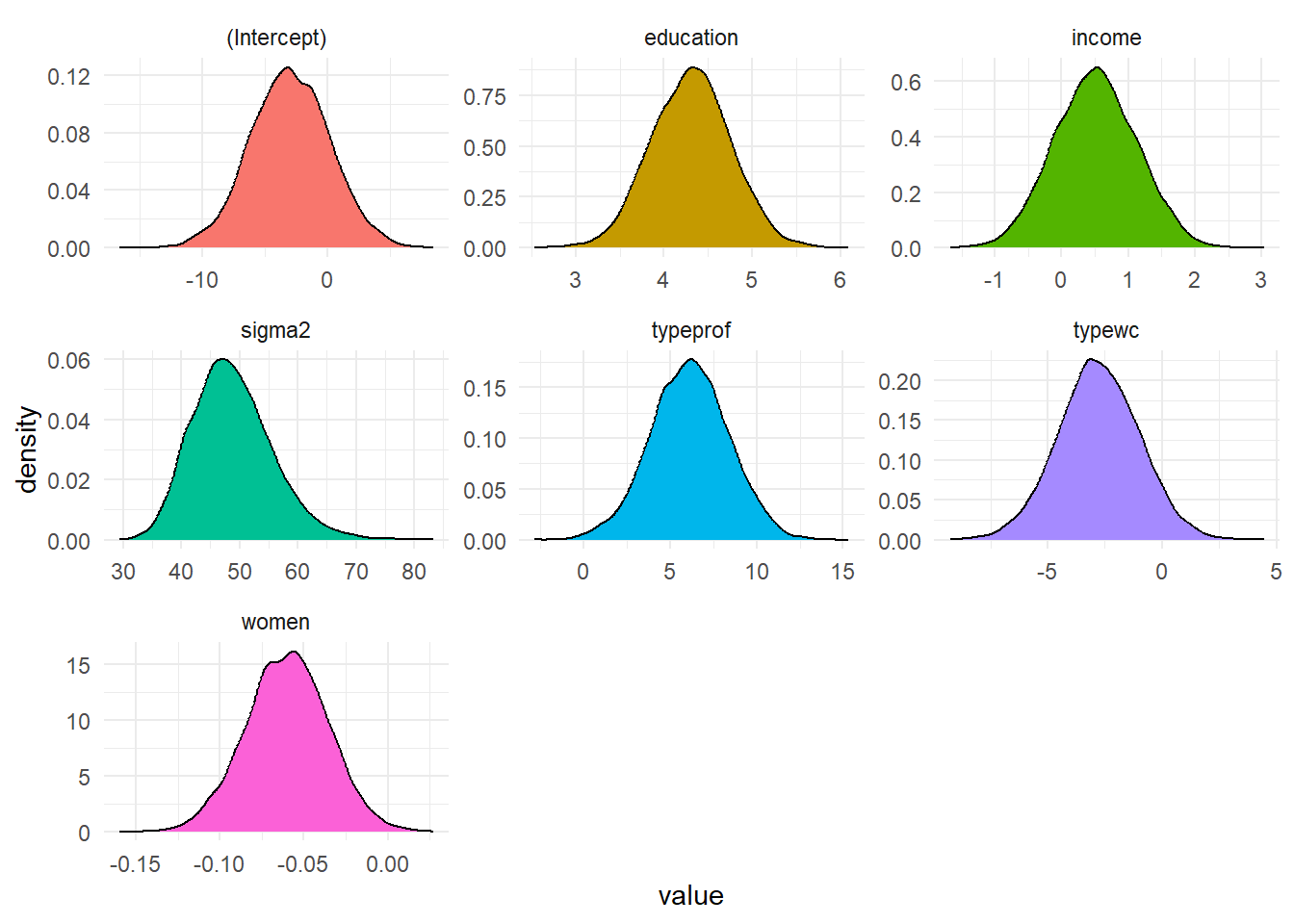
<- rstan:: extract (LM_stan, c ('beta' , 'sigma2' )) <- post_chain$ betacolnames (betas) <- colnames (X)<- post_chain$ sigma2<- data.frame (value = c (1 ], betas[,2 ], betas[,3 ],4 ], betas[,5 ], betas[,6 ],param = rep (c (colnames (X), 'sigma2' ), each = nrow (post_chain$ beta)))%>% ggplot (aes (value, fill = param)) + geom_density () + facet_wrap (~ param, scales = 'free' ) + theme_minimal () + theme (legend.position= "false" )
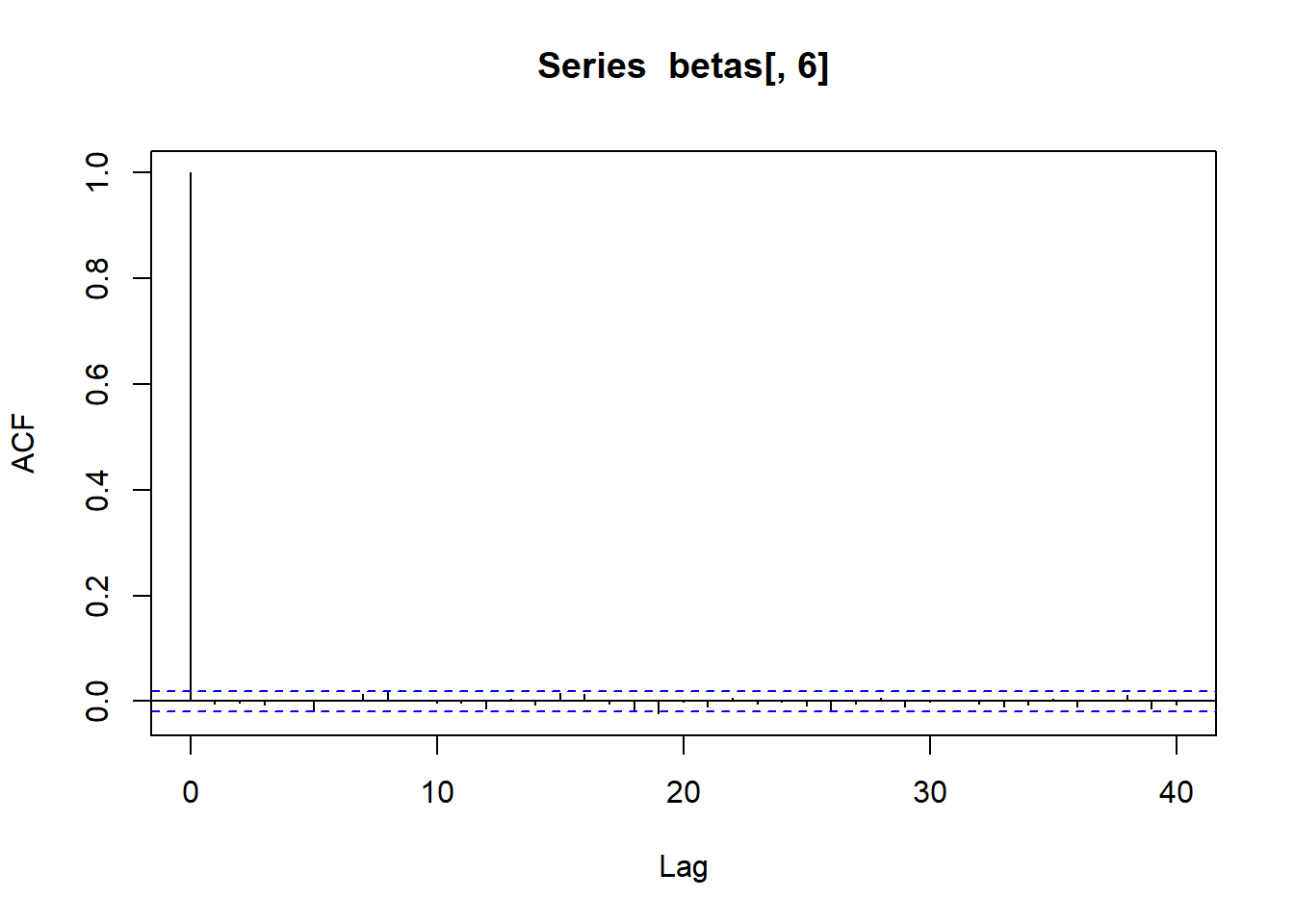
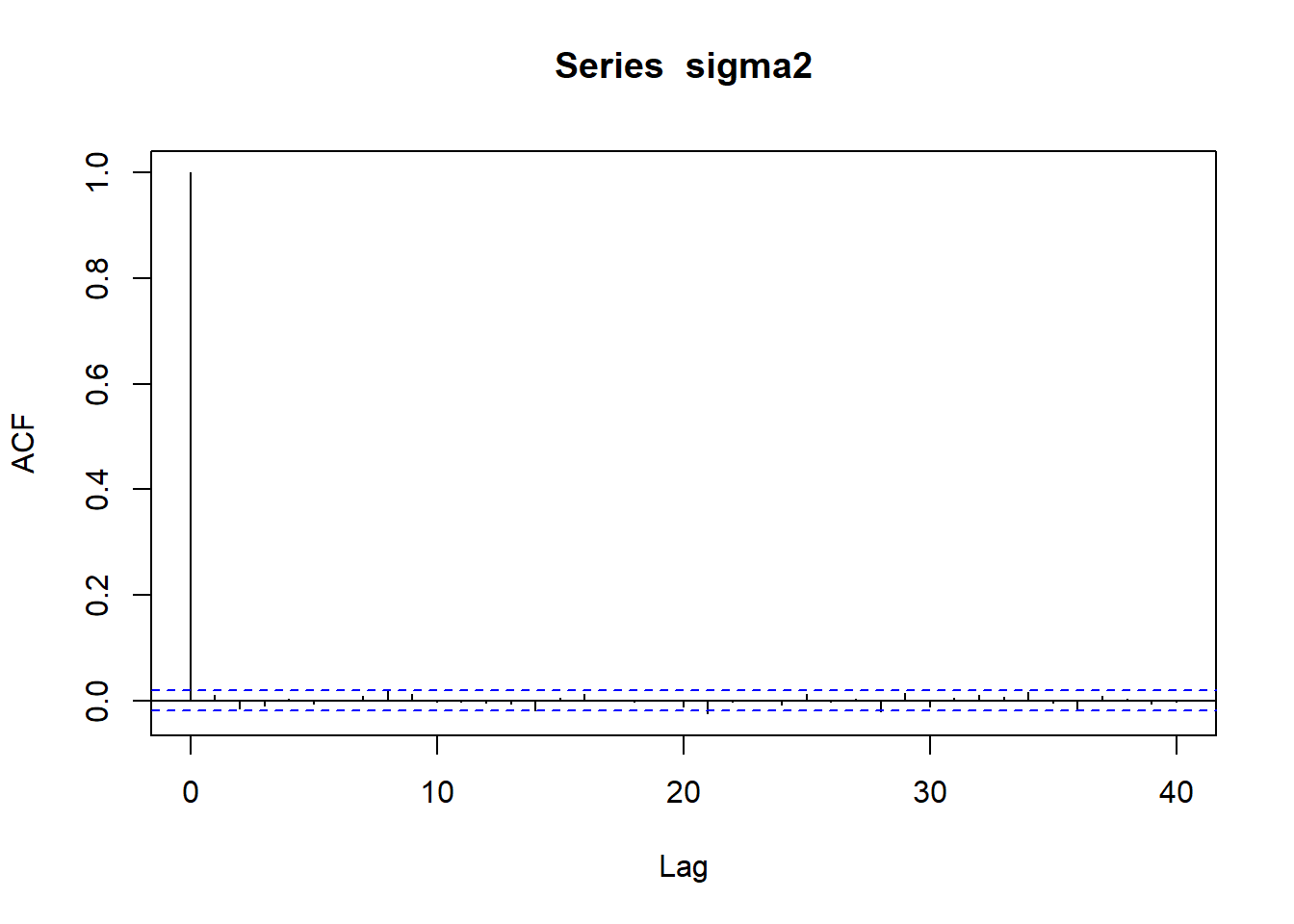
acf (betas[,5 ])acf (betas[,6 ])
As classical MCMCs, we can use the sampled values to approximate any quantity of interest. For example, we can approximate \(\mathbb{P}(\beta_1 > 4, \beta_3 > -0.5)\) :
mean (betas[,2 ] > 4 & betas[,4 ] > - .5 )
Logistic Regression
Let’s consider the logistic regression model to the cardiac dataset .
The Logistic_Model.stan model can be downloaded here .
rm (list = ls ())<- read.csv ("data/cardiac.csv" , header= T, sep= ";" )<- cardiac$ Chd<- model.matrix (lm (Chd ~ ., data= cardiac))<- nrow (X)<- ncol (X)
<- :: stan_model (file= "data/Logistic_Model.stan" )
S4 class stanmodel 'anon_model' coded as follows:
///////////////////////// DATA /////////////////////////////////////
data {
int<lower = 1> n; // number of data
int<lower = 1> K; // number of covariates (including the intercept)
int<lower = 0, upper = 1> y[n]; // response vector
matrix[n, K] X; // design matrix
vector[K] beta0;
vector[K] s2_0;
}
//////////////////// PARAMETERS /////////////////////////////////
parameters {
vector[K] beta; // regression coefficients
}
transformed parameters {
vector[n] eta;
vector<lower=0>[n] Odds;
vector<lower=0,upper=1>[n] p;
eta = X * beta;
for(i in 1:n){
Odds[i] = exp(eta[i]);
p[i] = Odds[i]/(1+Odds[i]);
}
}
////////////////// MODEL ////////////////////////
model {
// Prior
for (j in 1:K){
beta[j] ~ normal(beta0, sqrt(s2_0));
}
// Likelihood
for (s in 1:n){
y[s] ~ bernoulli(p[s]);
}
}
<- list (y = y,X = X,n = n,K = K,beta0 = rep (0 , K),s2_0 = rep (10 , K)
<- 10000 <- 2 <- rstan:: sampling (object = Logistic_Model,data = data.stan_logis,warmup = 0.5 * n.iter, iter = n.iter,thin = 1 , chains = nchain,refresh = n.iter/ 2
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 0.000102 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 1.02 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 10000 [ 0%] (Warmup)
Chain 1: Iteration: 5000 / 10000 [ 50%] (Warmup)
Chain 1: Iteration: 5001 / 10000 [ 50%] (Sampling)
Chain 1: Iteration: 10000 / 10000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 2.21 seconds (Warm-up)
Chain 1: 1.592 seconds (Sampling)
Chain 1: 3.802 seconds (Total)
Chain 1:
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 2).
Chain 2:
Chain 2: Gradient evaluation took 2.5e-05 seconds
Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.25 seconds.
Chain 2: Adjust your expectations accordingly!
Chain 2:
Chain 2:
Chain 2: Iteration: 1 / 10000 [ 0%] (Warmup)
Chain 2: Iteration: 5000 / 10000 [ 50%] (Warmup)
Chain 2: Iteration: 5001 / 10000 [ 50%] (Sampling)
Chain 2: Iteration: 10000 / 10000 [100%] (Sampling)
Chain 2:
Chain 2: Elapsed Time: 1.756 seconds (Warm-up)
Chain 2: 1.647 seconds (Sampling)
Chain 2: 3.403 seconds (Total)
Chain 2:
<- summary (Logistic_stan, pars = "beta" )$ summaryrownames (summ)[1 : K] <- colnames (X)round (summ, 3 )
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff
(Intercept) -4.431 0.021 0.952 -6.331 -5.064 -4.402 -3.776 -2.654 1974.032
Age 0.093 0.000 0.020 0.053 0.079 0.092 0.106 0.133 1973.055
Rhat
(Intercept) 1.002
Age 1.002
:: traceplot (Logistic_stan, pars = "beta" , inc_warmup = TRUE ):: traceplot (Logistic_stan, pars = "beta" , inc_warmup = FALSE )
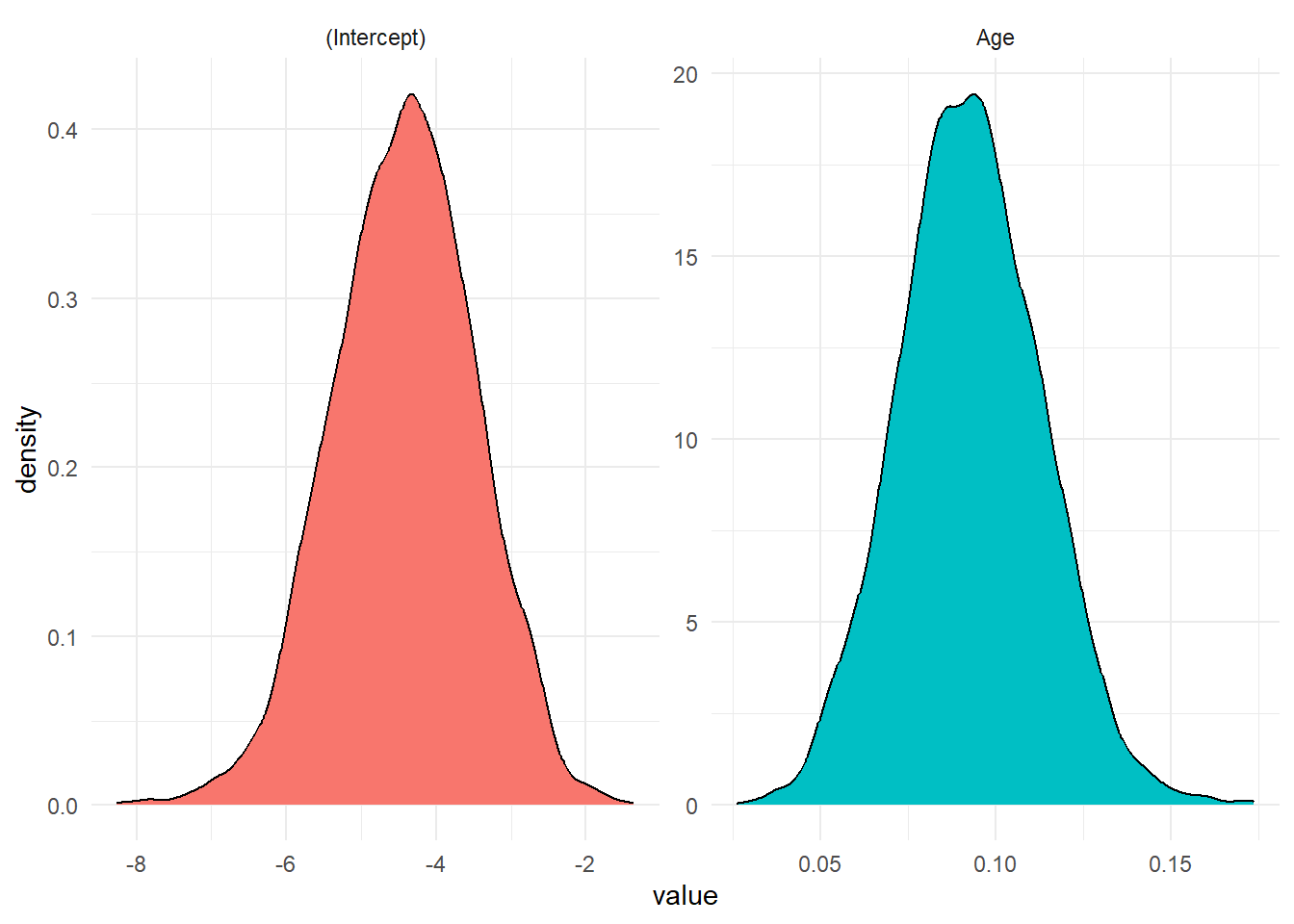
<- rstan:: extract (Logistic_stan, "beta" ) <- post_chain$ betacolnames (betas) <- colnames (X)<- data.frame (value = c (betas[,1 ], betas[,2 ]), param = rep (colnames (X), each = nrow (post_chain$ beta)))%>% ggplot (aes (value, fill = param)) + geom_density () + facet_wrap (~ param, scales = 'free' ) + theme_minimal () + theme (legend.position= "false" )
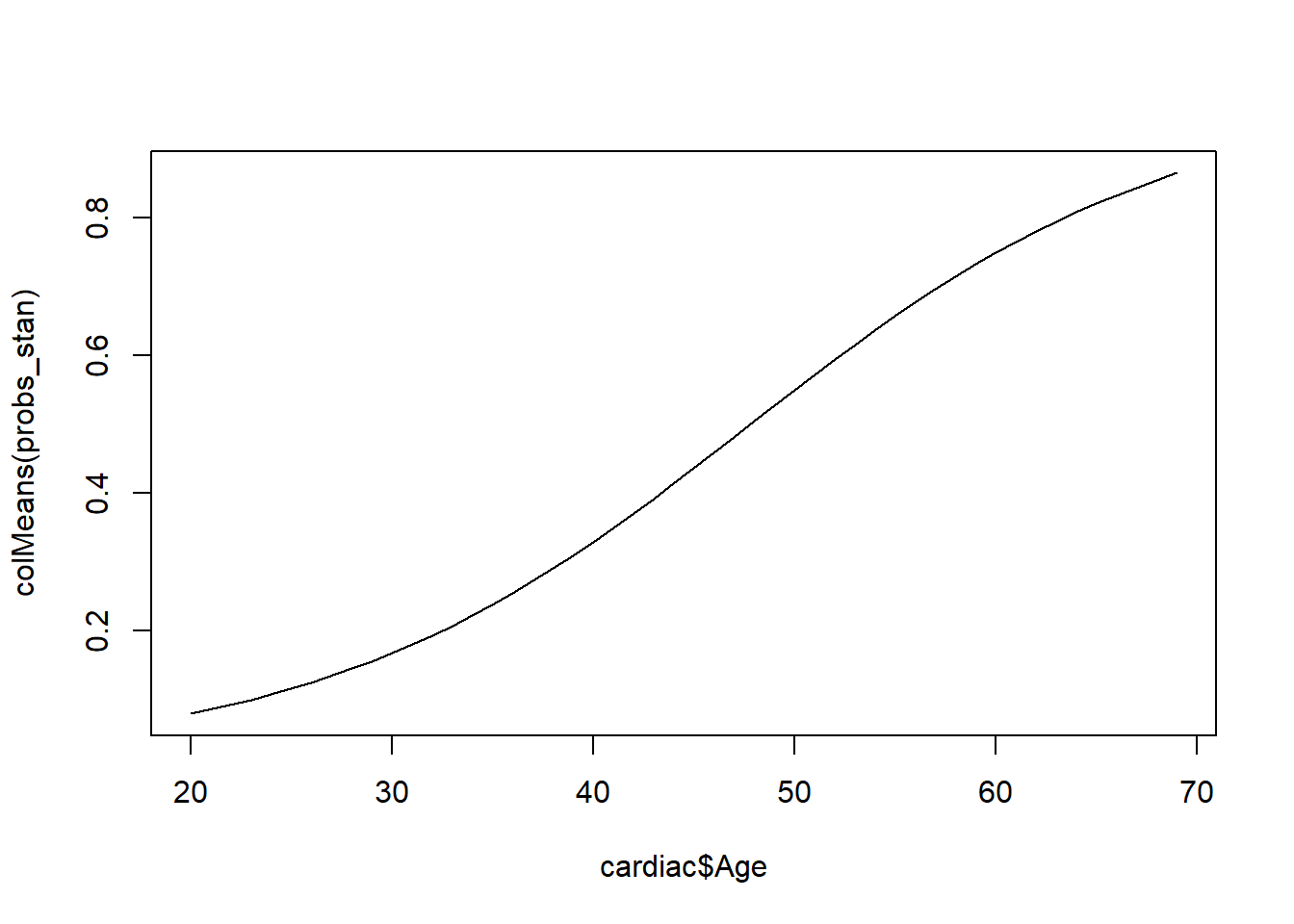
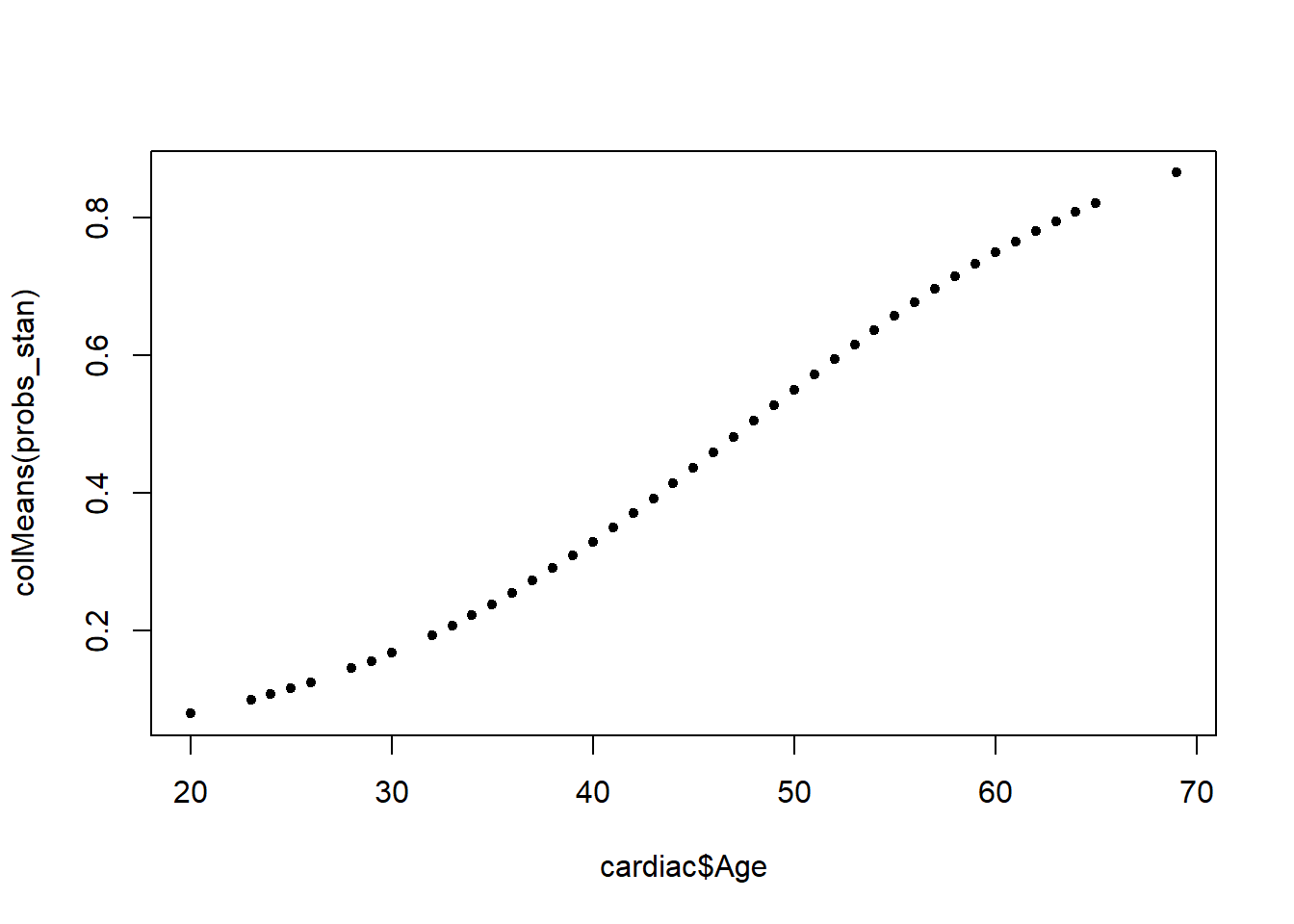
We can evaluate probabilities for each patients.
<- rstan:: extract (Logistic_stan, "p" )$ pplot (cardiac$ Age, colMeans (probs_stan), pch= 20 , type= "l" )plot (cardiac$ Age, colMeans (probs_stan), pch= 20 )